Combining Machine Learning and Molecular Dynamics to Predict P-Glycoprotein Substrates
Por um escritor misterioso
Descrição

Deep learning-based molecular dynamics simulation for structure-based drug design against SARS-CoV-2 - Computational and Structural Biotechnology Journal
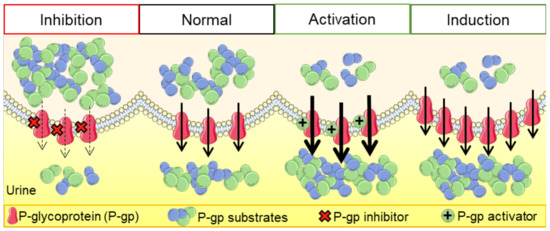
Molecules, Free Full-Text

Machine learning for small molecule drug discovery in academia and industry - ScienceDirect

P-glycoprotein Substrate Models Using Support Vector Machines Based on a Comprehensive Data set
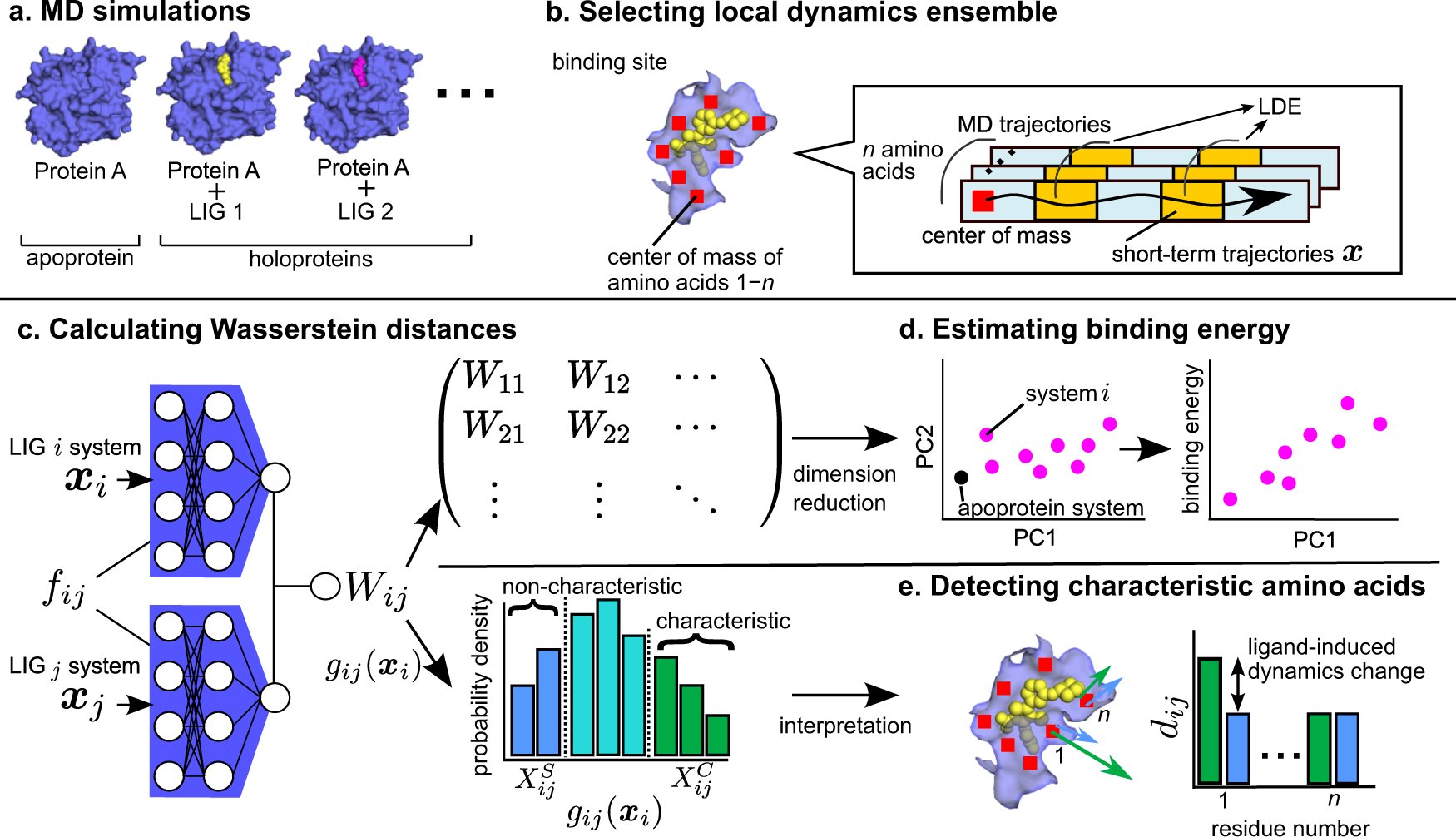
Differences in ligand-induced protein dynamics extracted from an unsupervised deep learning approach correlate with protein–ligand binding affinities

Kinetic modelling of the P-glycoprotein mediated efflux with a large-scale matched molecular pair analysis - ScienceDirect
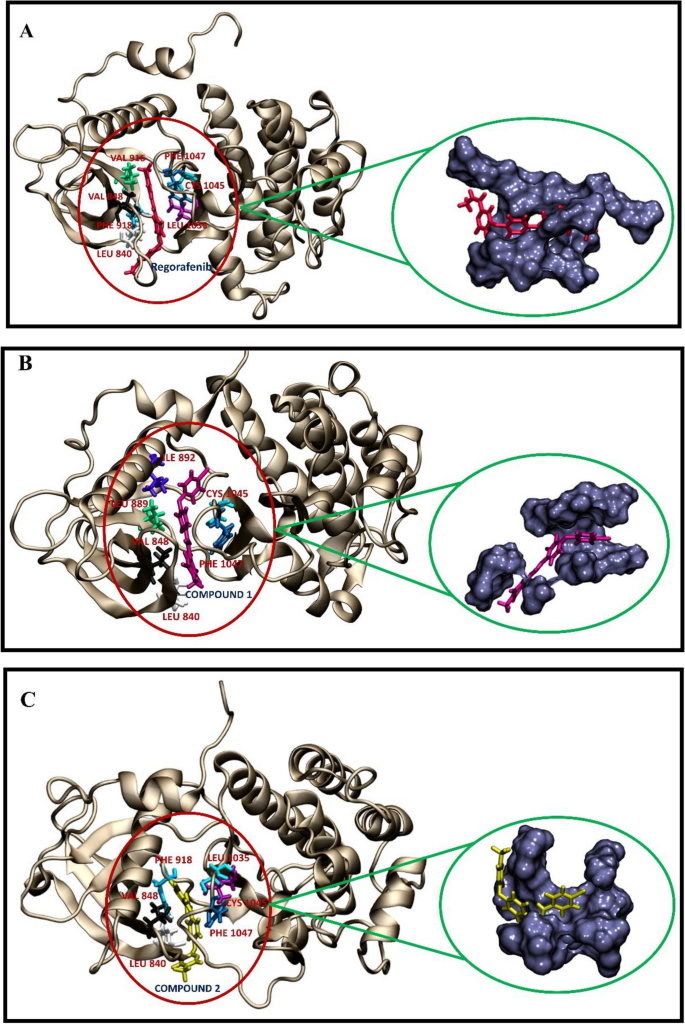
The use of machine learning modeling, virtual screening, molecular docking, and molecular dynamics simulations to identify potential VEGFR2 kinase inhibitors

Modelling peptide–protein complexes: docking, simulations and machine learning, QRB Discovery
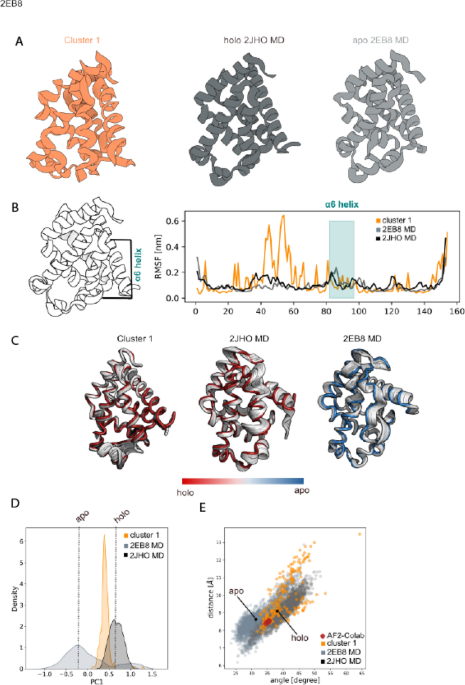
Machine learning/molecular dynamic protein structure prediction approach to investigate the protein conformational ensemble

Prediction and characterization of P-glycoprotein substrates potentially bound to different sites by emerging chemical pattern and hierarchical cluster analysis - ScienceDirect

Full article: Molecular docking, validation, dynamics simulations, and pharmacokinetic prediction of natural compounds against the SARS-CoV-2 main-protease
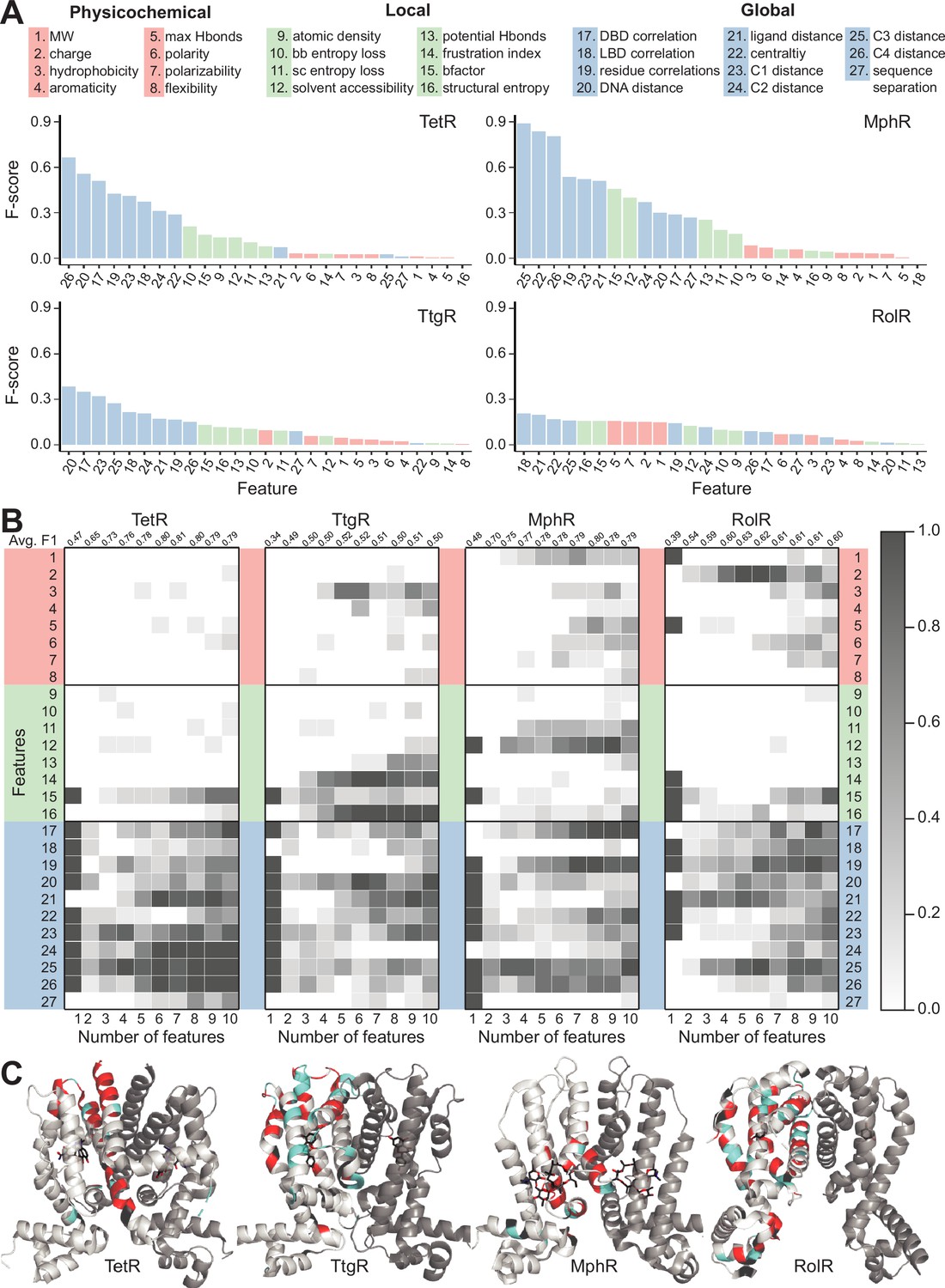
Deep mutational scanning and machine learning reveal structural and molecular rules governing allosteric hotspots in homologous proteins

Predicting drug metabolism and pharmacokinetics features of in-house compounds by a hybrid machine-learning model - ScienceDirect

Machine learning approaches and their applications in drug discovery and design - Priya - 2022 - Chemical Biology & Drug Design - Wiley Online Library
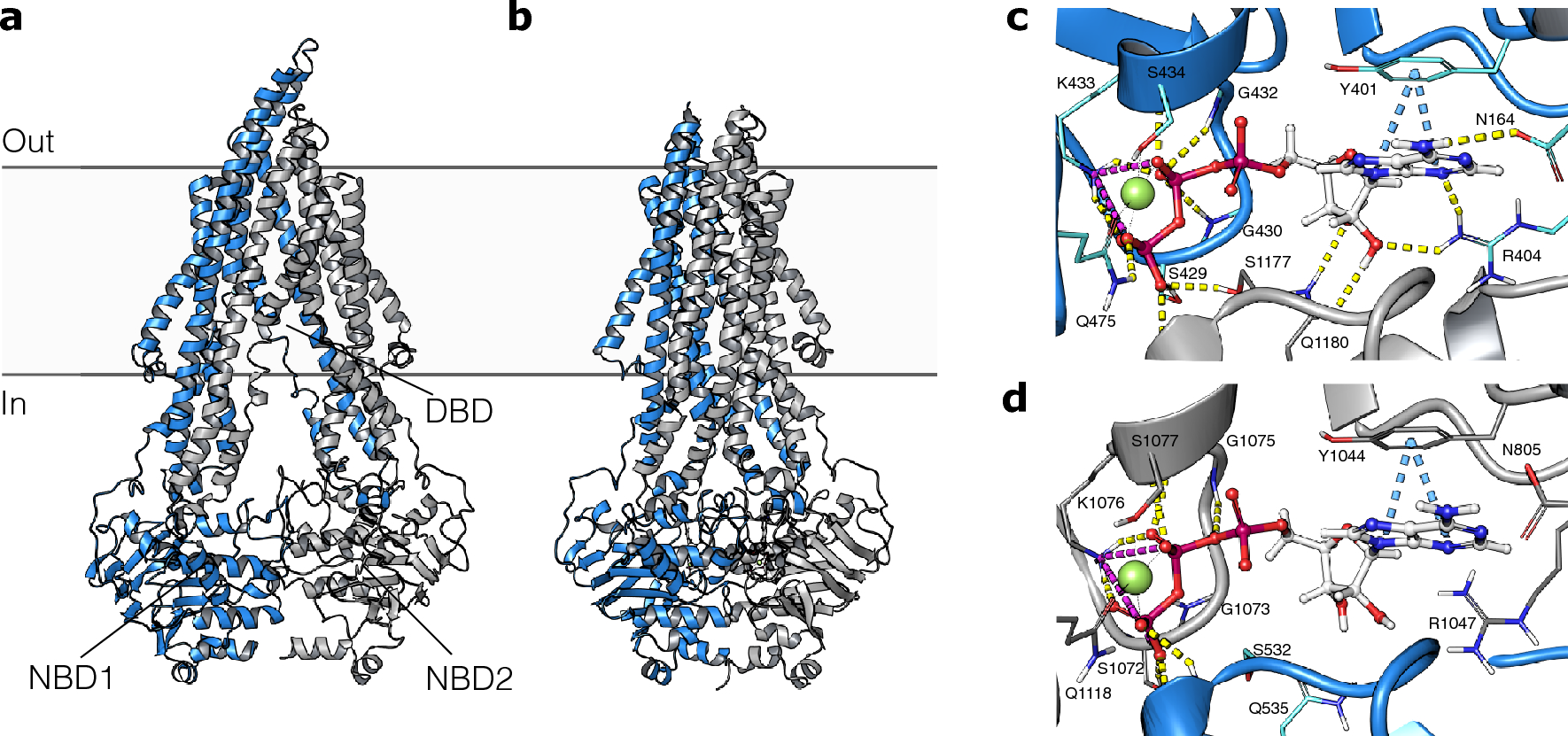
Structure-based discovery of novel P-glycoprotein inhibitors targeting the nucleotide binding domains
de
por adulto (o preço varia de acordo com o tamanho do grupo)